Signaling Lipidomics
Lipidomics.at turns virtual: ELM 2020 Bioinformatics for Lipidomics Symposium
Thanks to all attendees who helped to make this a great event! The ILS Applied Bioinformatics Interest Group invited to join the Bioinformatics for Lipidomics Symposium on October 2nd, 2020, as part of this year’s joint virtual European Lipidomics Meeting & Lipidomics Forum. With over 150 attendees discussing the challenges and future of lipidomics bioinformatics the event was a great success! We were very happy that this conference was supported by the ILS and the University of Vienna.
The schedule is available at ILS https://bit.ly/2IdlyJl
The symposium was divided into four major sessions:
Databases, Ontologies and Online Resources, Visualization and Exploration, Data Integration and Applications, Identification and Quantification Tools
The presentations of this symposium will be available soon via the ILS homepage!!
New Publication: Goslin
We introduce Goslin (August 2020), a polyglot grammar for common lipid shorthand nomenclatures based on the LipidMaps nomenclature and the shorthand nomenclature established by Liebisch et al. and used by LipidHome and SwissLipids. Goslin was designed to address the following pressing issues in the lipidomics field: 1) to simplify the implementation of lipid name handling for developers of mass spectrometry-based lipidomics tools; 2) to offer a tool that unifies and normalizes the main existing lipid name dialects enabling a lipidomics analysis in a high-throughput fashion. We provide implementations of Goslin in four major programming languages, namely C++, Java, Python 3, and R to kick-start adoption and integration. Further, we set up a web service for users to work with Goslin directly. All implementations are available free of charge under a permissive open source license. Here some links:
Lipidomics.at turns virtual: LipidCreator @ISMB
With great pleasure Lipidomics.at presented Lipid Creator on the annual meeting of the international Society for Computational Biology (ISCB) at the CompMS COSI track in Montreal (June 2020). Here comes the link: https://www.iscb.org/cms_addon/conferences/ismb2020/tracks/compmscosi
The lab also would like to thank all the people involved in the development of LIPIDCREATOR and the infrastructure around it and for their support! Also, thanks for the great questions and good suggestions during ISMB!
Lipidomics.at turns virtual: Virtual Podium
https://virtualpodium2020.wixsite.com/scienceandchilll
Social distancing is a must, but should not block the science! Due to COVID-19, you have been impacted by the cancellation of conferences and are losing the chance to connect and to present your research. Therefore the lipidomics.at team is embracing the opportunity to present at virtual platforms. Updates where we go and present can be found here, please join to engage and discuss. We are also hosting regularly own seminars on Thursdays, please contact us for more information.
Lipidomics.at turns virtual: Winter
Proteomics of the Lysosome
The lysosome is the central lytic organelle of mammalian cells. After it has been regarded for decades as a static and unregulated “cellular waste bag”, it is becoming more and more apparent, that the lysosome is a dynamic and regulated organelle, which is playing a central role in cellular metabolism. This is further underlined by the fact, that malfunctions of lysosomal hydrolases result in a group of >70 rare inherited diseases, so-called lysosomal storage disorders, and that lysosomes can play a decisive role in more common conditions such as cancer and neurodegenerative diseases. We are investigating the composition of lysosomes as well as the lysosomal response to various disease pathologies using mass spectrometry based proteomics. In particular, we optimized strategies for lysosomal enrichment, the proteomic characterization of lysosomal and lysosome-interacting proteins, and the identification of novel lysosomal proteins. Using Quantification Concatemers (QConCats), we generated absolutely quantified stable isotope labeled internal standards for the absolute quantification of lysosomal protein copy numbers and developed a multiple reaction monitoring (MRM) assay covering ~400 peptides from 143 proteins. Analysis of cell lines, isolated lysosomes, and mouse tissues revealed a dynamic range of several orders of magnitude for individual lysosomal proteins and organ-specific abundance profiles. For a better understanding of lysosomal protein-protein interactions and structures, we performed cross linking mass spectrometry experiments with isolated lysosomes. We were able to identify ~4300 cross links from >1300 proteins including 882 interactions for lysosomal and lysosome-associated proteins. This enabled us to identify novel interaction partners, to validate known structures, and to propose novel structures for lysosomal proteins and protein complexes. Taken together, these data provide us with a better view of lysosomal composition, providing valuable information for a better understanding of lysosomal function.
New Publication: Cellular stress causes cancer cell chemoresistance
Wien (UNIWIEN) – Resistance of cancer cells against therapeutic agents is a major cause of treatment failure, especially in recurrent diseases. An international team around the biochemists Robert Ahrends from the University of Vienna and Jan Medenbach from the University of Regensburg identified a novel mechanism of chemoresistance which has now been published in “Nature Communications”. It is driven by the Unfolded Protein Response, a cellular stress response pathway that alters gene expression and cellular metabolism to promote adaption and cell survival upon accumulation of unfolded proteins.
There is a broad range of mechanisms associated with chemoresistance, many of which to date are only poorly understood. The so-called cellular stress response – a set of genetic programmes that enable the cells to survive under stressful conditions – plays a key role in the development of numerous diseases and in chemoresistance. A better understanding of the cellular stress response pathways is therefore urgently required to develop new therapeutic concepts to overcome chemoresistance. “In this context, we employed comprehensive analytical approaches to gain deep and molecular insight into the Unfolded Protein Response, a cellular stress reaction induced by unfolded proteins”, says Robert Ahrends, group leader at the Department of Analytical Chemistry of the Faculty of Chemistry.
Unfolded proteins cause stress and disease
The Unfolded Protein Response (UPR) contributes to cancer development and progression and plays an important role in diseases such as diabetes and neurodegenerative disorders. For their study of the UPR’s molecular biological characteristics, the researchers applied state-to-art analytical tools in the context of a multiomics approach, combining large datasets from genetics, proteomics and metabolomics. This allowed them to define the Unfolded Protein Response regulon, a comprehensive list of genes that are activated to promote cell survival under stress.
“Besides the previously known factors, we identified to our surprise numerous genes that have not previously been implicated in stress response pathways”, explain the researchers, “and many of them have key functions in cancer development and cellular metabolism.”
Changes in 1C metabolism
Changes in cellular metabolism are characteristic of many cancer types and promote a rapid tumour growth, as Nobel Prize winner Otto Warburg demonstrated already in the 1930s in his ground-breaking work. In their study, the researchers discovered stress-mediated genetic regulation of enzymes involved in one-carbon (1C) metabolism which relies on the vitamin folate as a cofactor. Concomitant to the metabolic re-wiring, the stressed cells became fully resistant against chemotherapeutic agents, which target this specific metabolic pathway. This includes Methotrexate, a drug commonly employed in the treatment of cancer and rheumatic diseases. Detailed biochemical and genetic investigations revealed that resistance is driven by a previously unrecognized mechanism. According to the study authors, its precise molecular characterisation might lead to novel therapeutic concepts aimed at overcoming chemoresistance in cancer therapy.
Publikation in “Nature Communications”:
Reich S, Nguyen CDL, Has C, Steltgens S, Soni H, Coman C, Freyberg M, Bichler A, Seifert N, Conrad D, Knobbe-Thomsen CB, Tews B, Toedt G, Ahrends R, und Medenbach J: A multi-omics analysis reveals the Unfolded Protein Response regulon and a role of eIF2-phosphorylation in resistance to folate-based anti-metabolites.
DOI: 10.1038/s41467-020-16747-y
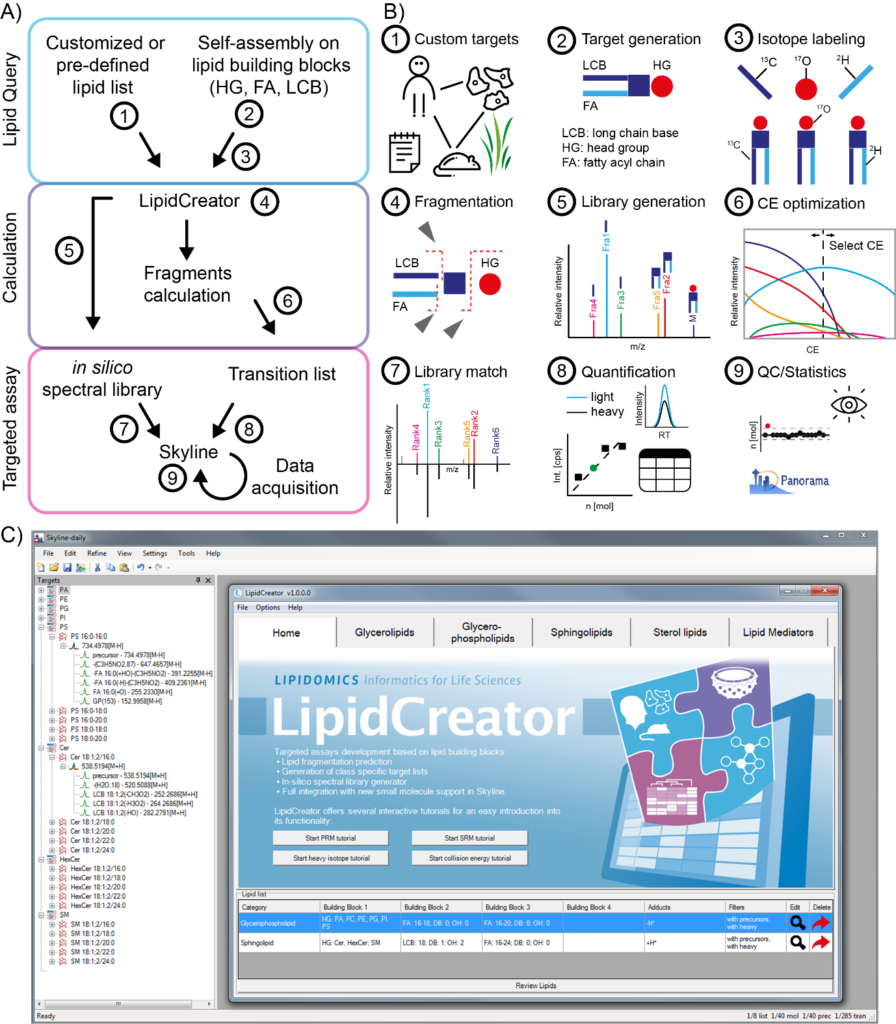
New Publication: LipidCreator
Mass spectrometry (MS)-based targeted lipidomics enables the robust quantification of selected lipids under various biological conditions but comprehensive software tools to support such analyses are lacking. Here we present LipidCreator (LC), a software that fully supports targeted lipidomics assay development. LC offers a comprehensive framework to compute MS/MS fragment masses for over 60 lipid classes. LC provides all functionalities needed to define fragments, manage stable isotope labeling, optimize collision energy and generate in silico spectral libraries. We validate LC assays computationally and analytically and prove that it is capable to generate large targeted experiments to analyze blood and to dissect lipid-signaling pathways such as in human platelets.
Lipidomics.at turns virtual: Witting

Helmhotz Zentrum Muenchen
This week we are looking forward to a presentation from Michael Witting from the Helmholtz Zentrum München. Michael is especially interested in lipidomics and metabolomics of C. elegans and we are anticipating his talk about LC-MS based strategies for the study of this popular model organism. The talk will take place on Thursday, 10 a.m. via Zoom. Registration is still possible, just write an e-mail to robert.ahrends@univie.ac.at. Thanks Michael for joining!