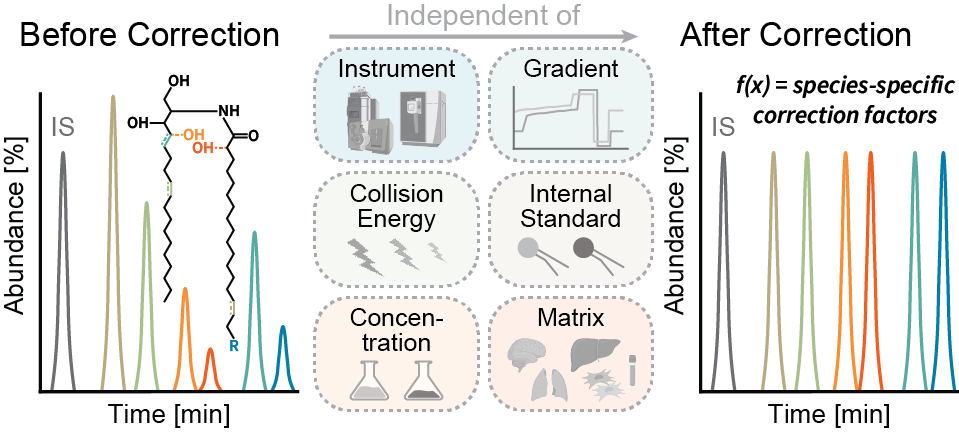
Accurate quantification of molecular sphingolipid species is essential for a comprehensive understanding of their diverse roles in physiological and pathological contexts. The conventional method of quantifying sphingolipids, which relies on sphingoid base-derived fragments relative to a class-specific internal standard, often falls short in addressing the structural diversity of these bioactive molecules. To overcome this limitation, we have developed a novel fragmentation model to correct for structural differences, providing a solution that transcends the constraints of the traditional “one standard per class” strategy. Notably, our approach is independent of the internal standard, instrumental setup, and collision energy. Furthermore, we have integrated this correction method into a user-friendly KNIME workflow. Validation results affirm the efficacy of our approach in accurately quantifying ceramide subclasses across diverse biological matrices. This advancement opens new horizons for exploring sphingolipid metabolism, offering profound insights into its implications.
Go to the publication here.

