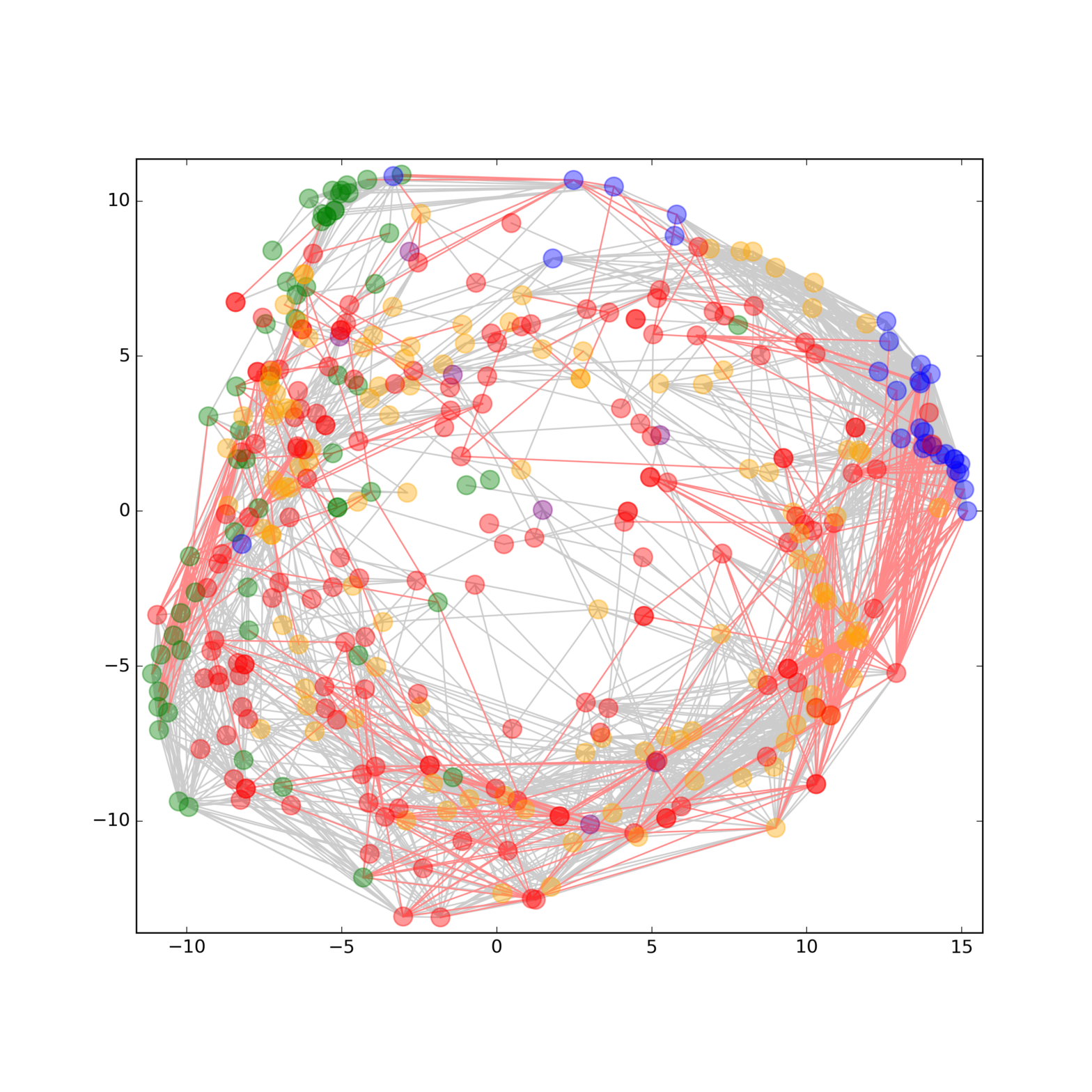
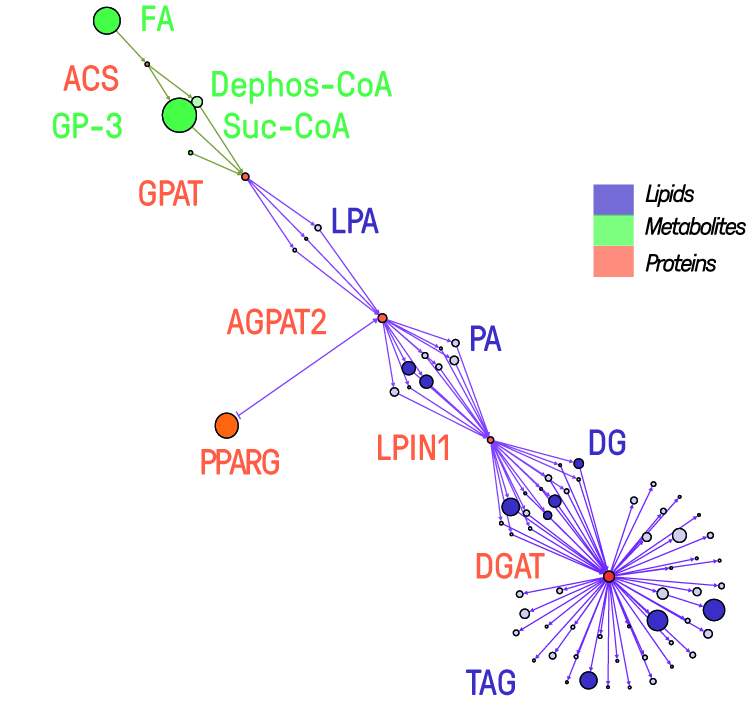
 The rising prevalence of obesity globally presents a critical social challenge, threatening to reverse the progress made in life expectancy in developed nations. Gaining a thorough understanding of the mechanisms behind fat cell production is, thus, essential for the development of novel treatment strategies. While past studies focused on individual molecular layers, we recognize the importance of considering higher network connectivity levels, particularly lipid feedback controlling the master regulator of adipogenesis, PPARG. Our approach integrates advanced lipidomics and proteomics techniques, monitoring PPARG and the lipidome during perturbations in vitro. Through innovative multiomics strategies and high-throughput imaging, we dissect feedback networks and identify lipid regulators of PPARG. This research not only advances scientific knowledge but also holds promise for future obesity drug development, potentially revolutionizing treatment approaches.
The rising prevalence of obesity globally presents a critical social challenge, threatening to reverse the progress made in life expectancy in developed nations. Gaining a thorough understanding of the mechanisms behind fat cell production is, thus, essential for the development of novel treatment strategies. While past studies focused on individual molecular layers, we recognize the importance of considering higher network connectivity levels, particularly lipid feedback controlling the master regulator of adipogenesis, PPARG. Our approach integrates advanced lipidomics and proteomics techniques, monitoring PPARG and the lipidome during perturbations in vitro. Through innovative multiomics strategies and high-throughput imaging, we dissect feedback networks and identify lipid regulators of PPARG. This research not only advances scientific knowledge but also holds promise for future obesity drug development, potentially revolutionizing treatment approaches. This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.