Turning hay into needles: the road to integration of large-scale analytical lipidomics data into biomedical research
Lipidomics studies networks, pathways and interactions of cellular lipids within the biological system.
As a newly emerging discipline and highly
interdisciplinary field based on the application of mass spectrometry,
analytical chemistry and computational biology, lipidomics is evolving into a
promising stepping stone for systems biology research with recent applications
in human health and disease.
A rapid increase in the number of measurements,
as well as increasing amounts of data per measurement due to higher
sensitivity, generate massive data sets that pose a challenge to traditional
data analysis and integration.
Similar to other “omics” disciplines,
lipidomics is thus facing the “big data trap”, challenging the ways we store
and process large amounts of data acquired over time, extracting useful
information from it, and moreover, interpreting the data from a systems
medicine research perspective.
The bioinformatics team of our lab works on
projects (https://lifs.isas.de/) to
overcome these challenges and develops computational tools and programs
covering the requirements of life science researchers. These tools are
first tested internally by our team members who generate the data, and are
subsequently optimized and released for general use to the scientific
community. Moreover, we also organize and arrange workshops, where our team
teaches how to use and implement our programs and tools, helping the user to
achieve scientifically meaningful data from their research.
To advertise the newest generation of lipid analysis tools and programs, we hosted the LipoSysMed summer school 2019 in alliance with the research groups of Dr. Maria Fedorova (Institute of Bioanalytical Chemistry) and Dr. Robert Ahrends (Institut für Analytische Wissenschaften – ISAS) in Leipzig / Germany (https://home.uni-leipzig.de/liposysmed/) . For one week, the participants got in touch with the newest computational technologies and attended workshops and tutorials. The rationale for the summer school was to bring practitioners with diverse clinical research backgrounds together to network with the bioinformatics tool developers and their colleagues. At LipoSysMed, we had the opportunity for mutual exchange and to troubleshoot scientific challenges with the tool developers directly, giving us fresh ideas on application-oriented targets and providing us with even more input on user-friendly software development. Overall, we especially set a high value on the constant bilateral communication with clinical staff scientists and tool developers, to increase the usability and functionality of our analysis programs, in order to reduce analysis time and getting most out of very costly clinical samples.
Our message is that just collecting “big data” is
meaningless without proper analytics. We are convinced that only with
consecutive and routine teaching and support of our fellow researchers, an
increasing number of laboratories will introduce and extend their
bioinformatics pipelines for the benefit of more insightful, reproducible
research.
We design next-generation lipid analysis tools
for tailored analytics, enhancing and bringing forward research on topics such
as the development of Obesity, Alzheimer disease or insufficiency in blood
clotting, which are all highly lipid-dependent.
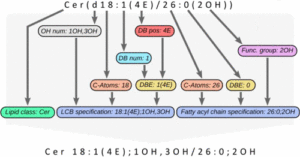 The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source. https://pubs.acs.org/doi/10.1021/acs.analchem.1c05430
The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source. https://pubs.acs.org/doi/10.1021/acs.analchem.1c05430