9th European Workshop on Lipid Mediators
ASMS 2024
A Targeted, Bioinert LC–MS/MS Method for Sensitive, Comprehensive Analysis of Signaling Lipids
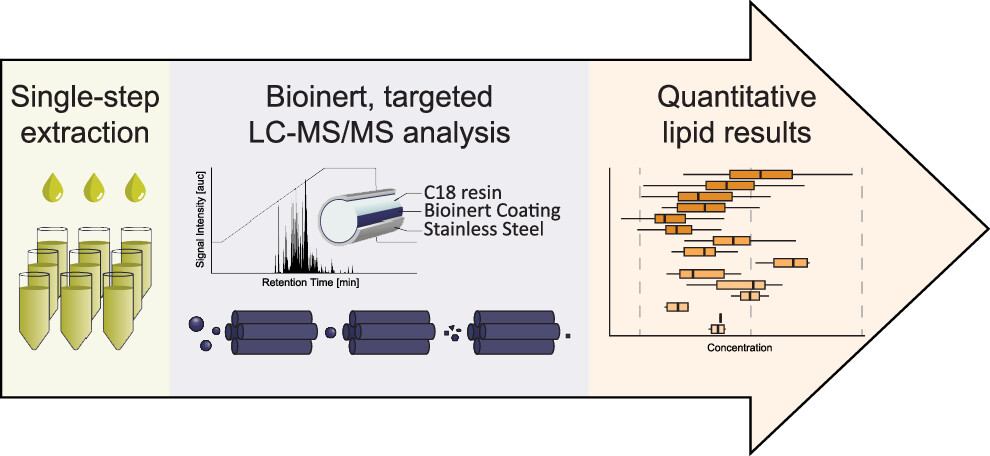
A new publication of Stefanie is here!
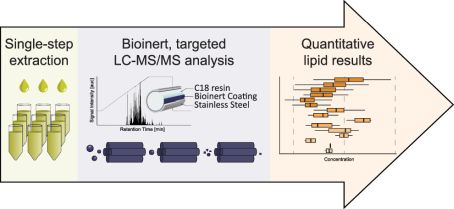
Signaling lipids play crucial roles in how cells function. However, it’s been difficult to monitor all these lipids at once because of their complexity and their chemical nature. We’ve developed a new, sensitive method using liquid chromatography-mass spectrometry (LC-MS/MS) that can accurately track a wide range of these lipids. Our method involves a straightforward sample extraction and uses specialized equipment to analyze 17 different classes of signaling lipids. This process is highly sensitive and consistent, allowing us to monitor 388 lipids in just 20 minutes. We tested our method on human plasma and successfully measured 307 different lipid molecules. We also studied changes in lipid signaling during platelet activation, finding many important regulatory pathways. This new technique opens up the possibility for detailed and comprehensive analysis of signaling lipids in complex biological systems.
Go to this publication!
Rubenzucker S, Manke MC, Lehmann R, Assinger A, Borst O, Ahrends R. A Targeted, Bioinert LC-MS/MS Method for Sensitive, Comprehensive Analysis of Signaling Lipids. Anal Chem. 2024 May 25. doi: 10.1021/acs.analchem.4c01388. Epub ahead of print. PMID: 38795073.









 The rising prevalence of obesity globally presents a critical social challenge, threatening to reverse the progress made in life expectancy in developed nations. Gaining a thorough understanding of the mechanisms behind fat cell production is, thus, essential for the development of novel treatment strategies. While past studies focused on individual molecular layers, we recognize the importance of considering higher network connectivity levels, particularly lipid feedback controlling the master regulator of adipogenesis, PPARG. Our approach integrates advanced lipidomics and proteomics techniques, monitoring PPARG and the lipidome during perturbations in vitro. Through innovative multiomics strategies and high-throughput imaging, we dissect feedback networks and identify lipid regulators of PPARG. This research not only advances scientific knowledge but also holds promise for future obesity drug development, potentially revolutionizing treatment approaches.
The rising prevalence of obesity globally presents a critical social challenge, threatening to reverse the progress made in life expectancy in developed nations. Gaining a thorough understanding of the mechanisms behind fat cell production is, thus, essential for the development of novel treatment strategies. While past studies focused on individual molecular layers, we recognize the importance of considering higher network connectivity levels, particularly lipid feedback controlling the master regulator of adipogenesis, PPARG. Our approach integrates advanced lipidomics and proteomics techniques, monitoring PPARG and the lipidome during perturbations in vitro. Through innovative multiomics strategies and high-throughput imaging, we dissect feedback networks and identify lipid regulators of PPARG. This research not only advances scientific knowledge but also holds promise for future obesity drug development, potentially revolutionizing treatment approaches.