This website uses cookies so that we can provide you with the best user experience possible. Cookie information is stored in your browser and performs functions such as recognising you when you return to our website and helping our team to understand which sections of the website you find most interesting and useful.
LipidSpace LIFS Tool
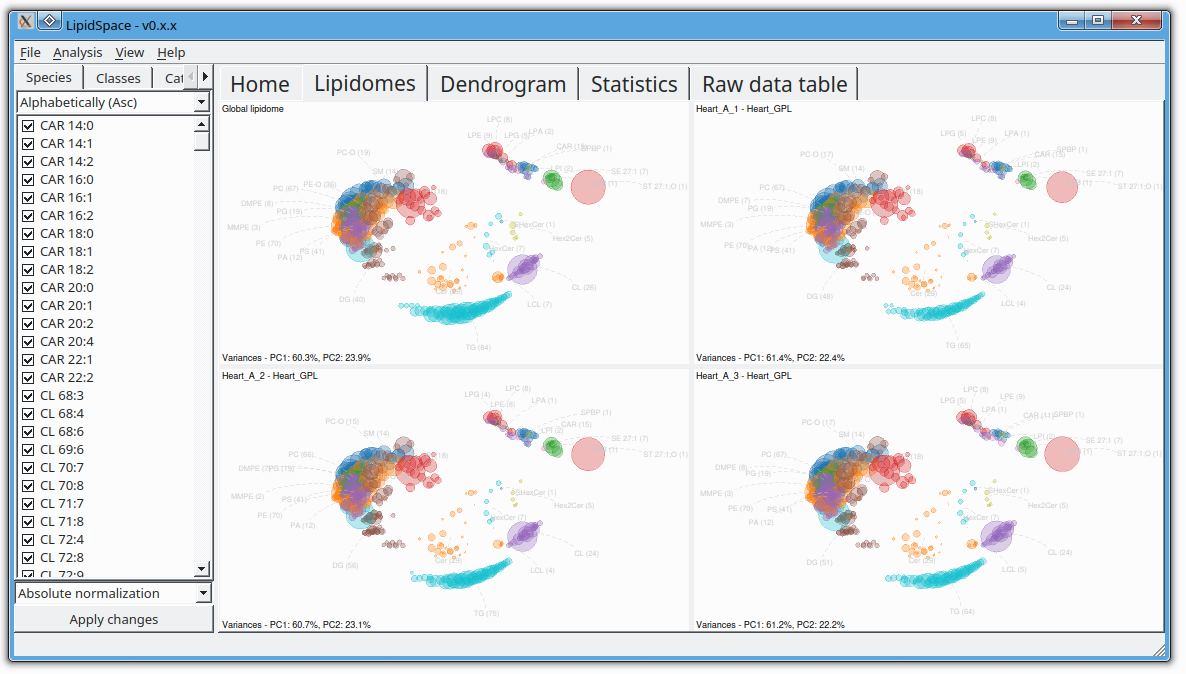
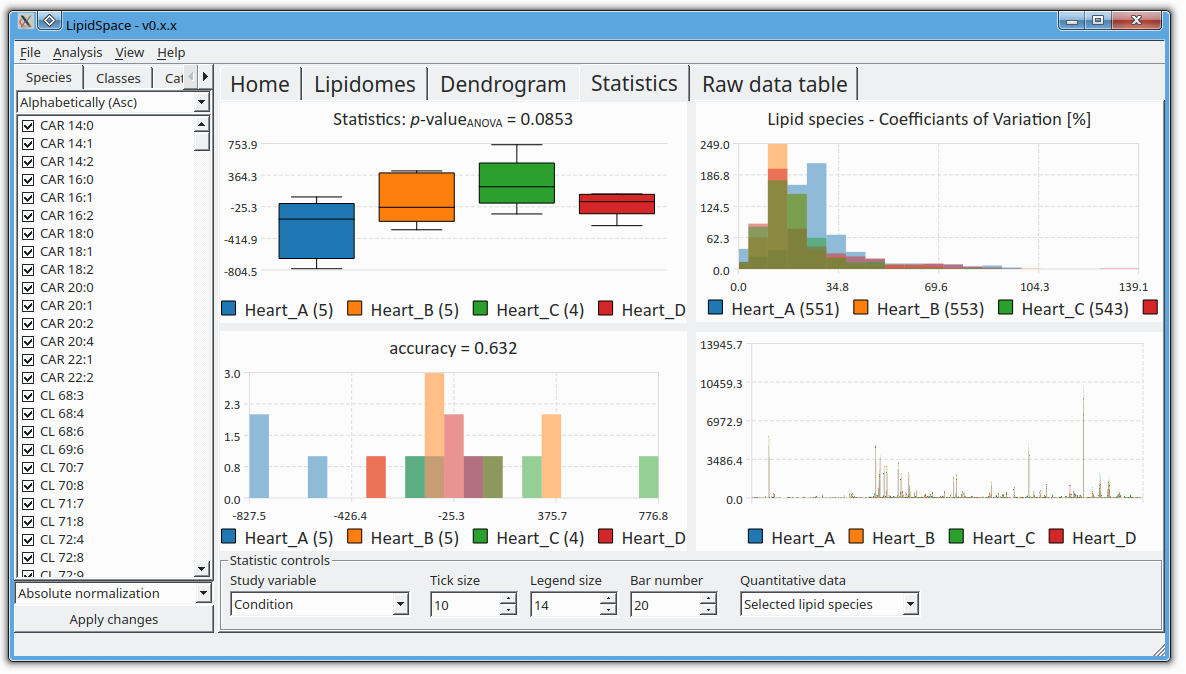
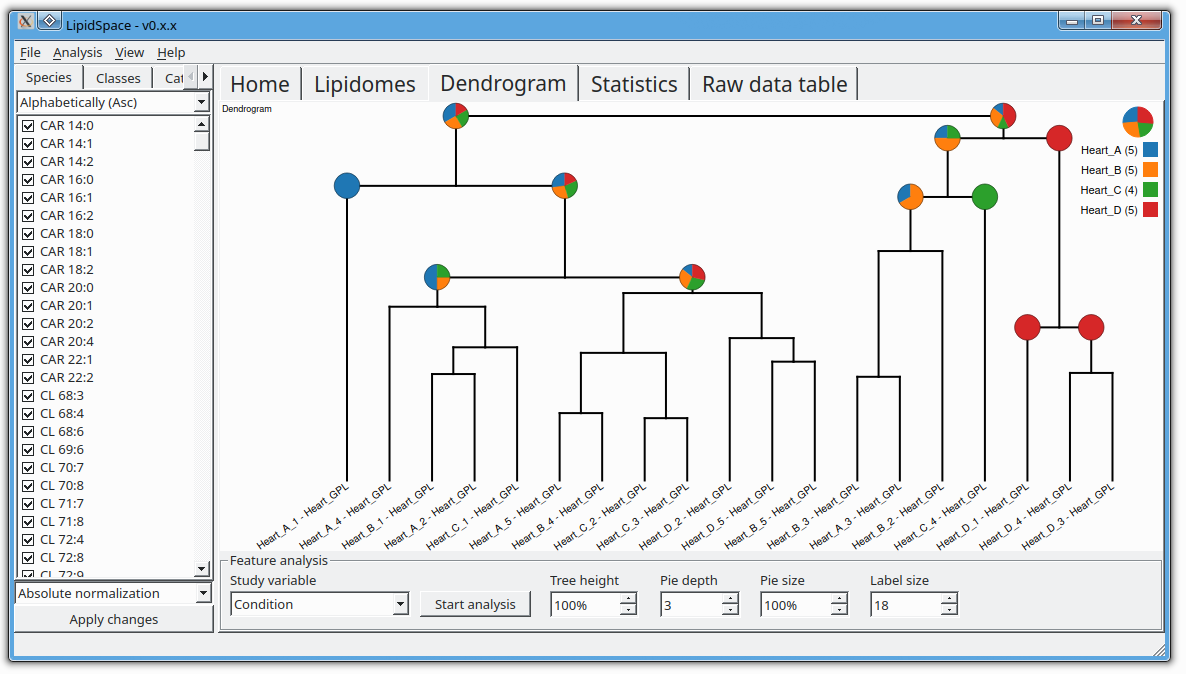
LipidSpace is a stand-alone tool to analyze and compare lipidomes by assessing their structural differences.
A graph-based comparison of lipid structures that allows to calculate distances between lipids and to determine similarities across lipidomes. It allows for a rapid (re)analysis of experiments, identifies lipids responsible for shaping the respective lipidome, and provides methods for quality control.
LipidSpace has been built and tested under Windows 10 and Ubuntu 22.04 Linux. It comes with four built-in tutorials to get you started.
More: https://lifs-tools.org/lipidspace.html
Computational Lipidomics
Platelet Lipidomics
Platelet integrity and function critically depend on lipid composition. However, the lipid inventory in platelets was hitherto not quantified (Peng et al., Blood, 2018). Today our lab examines the lipidome of murine and human platelets using lipid-category tailored protocols on quantitative lipidomics platforms. We commonly can cover the platelet lipidome, which is comprised out of 500 lipids (99.9% of the total lipid mass) over a concentration range of seven orders of magnitude. We conduct systematic comparison of lipidomics network in resting and activated murine platelets, validated in human platelets, where we inter alia revealed that less than 20% of the platelet lipidome is changed upon activation, involving mainly lipids containing arachidonic acid. However, the most interesting work that we currently conducting in close collaboration with our partners is the analysis of different diseases models (Scheller et al.,Haematologica, 2019) which display and thrombotic phenotype. E.g., Sphingomyelin phosphodiesterase-1 (Smpd1) deficiency results in a very specific modulation of the platelet lipidome (Peng et al., Blood, 2018) with an order of magnitude up-regulation of lyso-sphingomyelin (SPC), and subsequent modification of platelet activation and thrombus formation, which sheds light on novel mechanisms important for platelet function, and has therefore the potential to open novel diagnostic and therapeutic opportunities.




