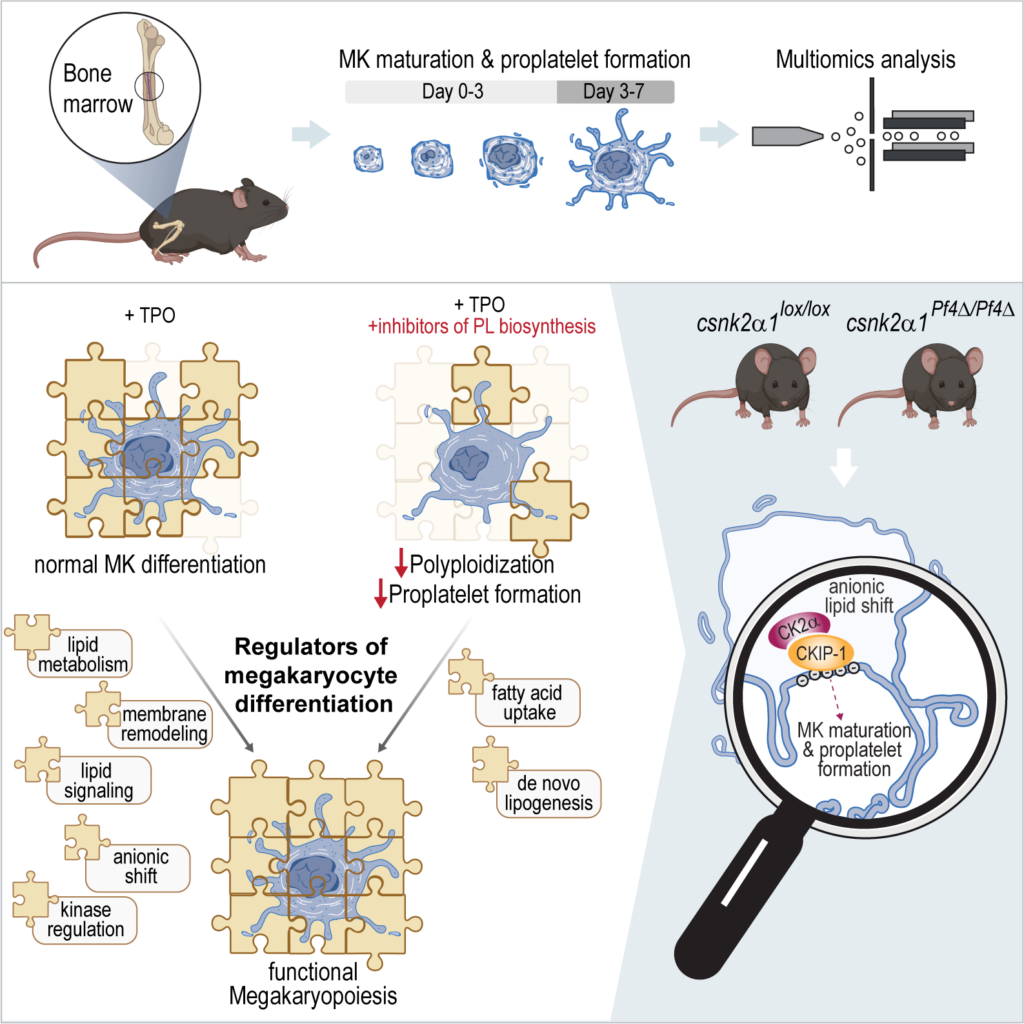
Scientists unveiled a deeper understanding of megakaryocyte differentiation and blood platelet production, a process crucial for maintaining healthy blood clotting and preventing excessive bleeding. The study featured in “Nature Cardiovascular Research,” led by chemist Robert Ahrends from the University of Vienna and cardiologist Oliver Borst from the University of Tübingen, sheds light on the intricate role of lipids – the building blocks of cell membranes – in the formation of these vital blood components.
Blood platelets, tiny disk-shaped cells in our bloodstream, play a pivotal role in wound healing and preventing excessive bleeding. The process by which these platelets are produced, known as thrombopoiesis, begins with the differentiation of specialized cells called megakaryocytes. These megakaryocytes undergo a series of transformations, ultimately giving rise to thousands of blood platelets.
“It is astonishing that despite its clinical importance, neither a quantitative lipid inventory nor a map of the lipid metabolism of megakaryocytes has been available,” says Robert Ahrends.
So far, the study of megakaryocyte differentiation and platelet production has centered on enzymes related only to sphingolipid metabolism, but until now, the exact nature of the lipid species involved, as well as their metabolizing enzymes and their impact on the process, remained largely unknown. Using advanced mass spectrometry-based multiomics, the research team investigated the mechanisms of lipidome modulation of megakaryocytes as they matured and formed platelets.
Shaping anionic membranes: A lipid-driven transformation
As the megakaryocytes differentiated, they underwent substantial changes in lipid membrane composition, resulting in what the team termed an “anionic membrane phenotype.” This shift was found to be directly linked to the regulation of crucial proteins and kinases involved in platelet formation.
The study also highlighted the role of fatty acids – the building blocks of lipids – in this process. The researchers found that the uptake of fatty acids increased significantly during megakaryocyte maturation, alongside a boost in fatty acid synthesis. Manipulation of such resulted in profound thrombocytopenia, a condition characterized by abnormally low platelet levels.
Importance for cardiovascular fitness
In essence, this study offers a remarkable new perspective on megakaryocyte differentiation and platelet production, highlighting the interplay between lipids and proteins on cellular membrane transformation. “Our study provides a foundation for exploring how disruptions in lipid metabolism, seen in conditions like obesity, might affect platelet production and overall cardiovascular health,” says cardiologist Oliver Borst. Beyond scientific insights, this discovery holds promise for new avenues in addressing health challenges on the horizon.
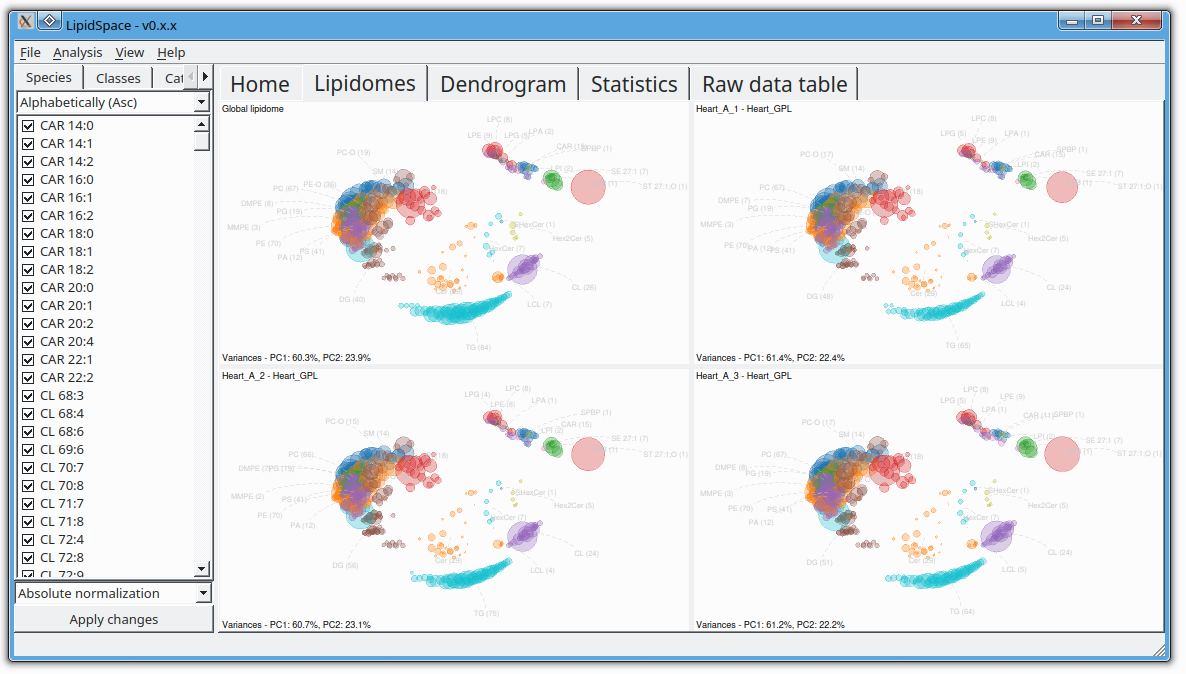
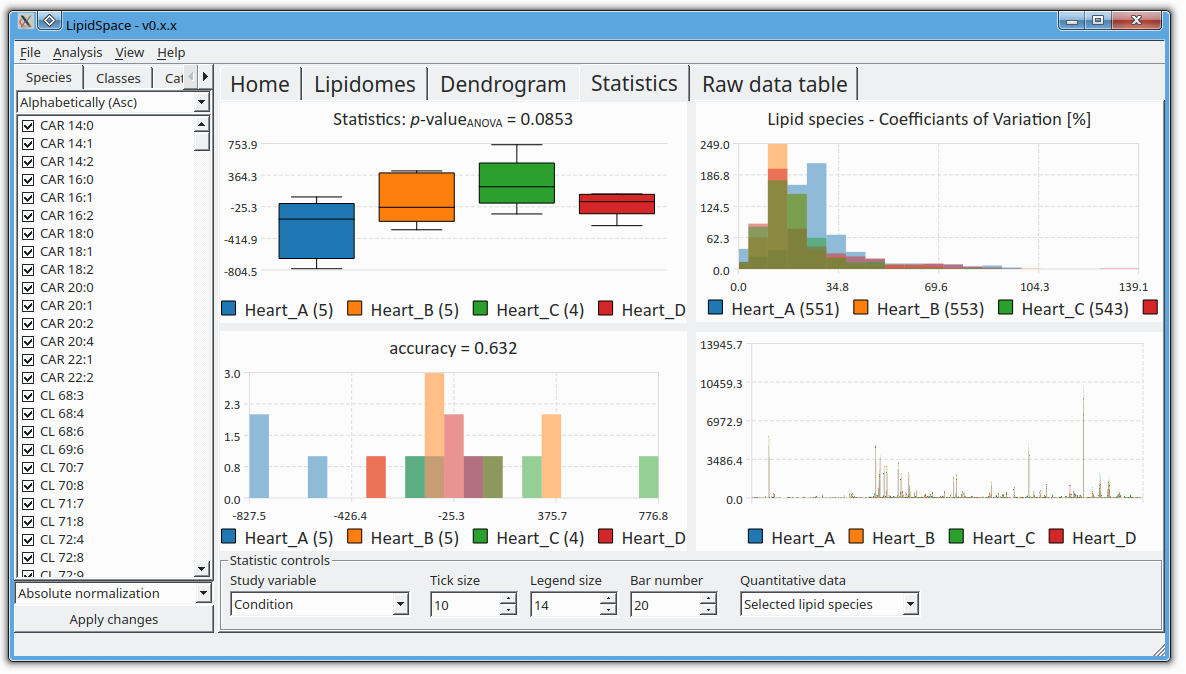
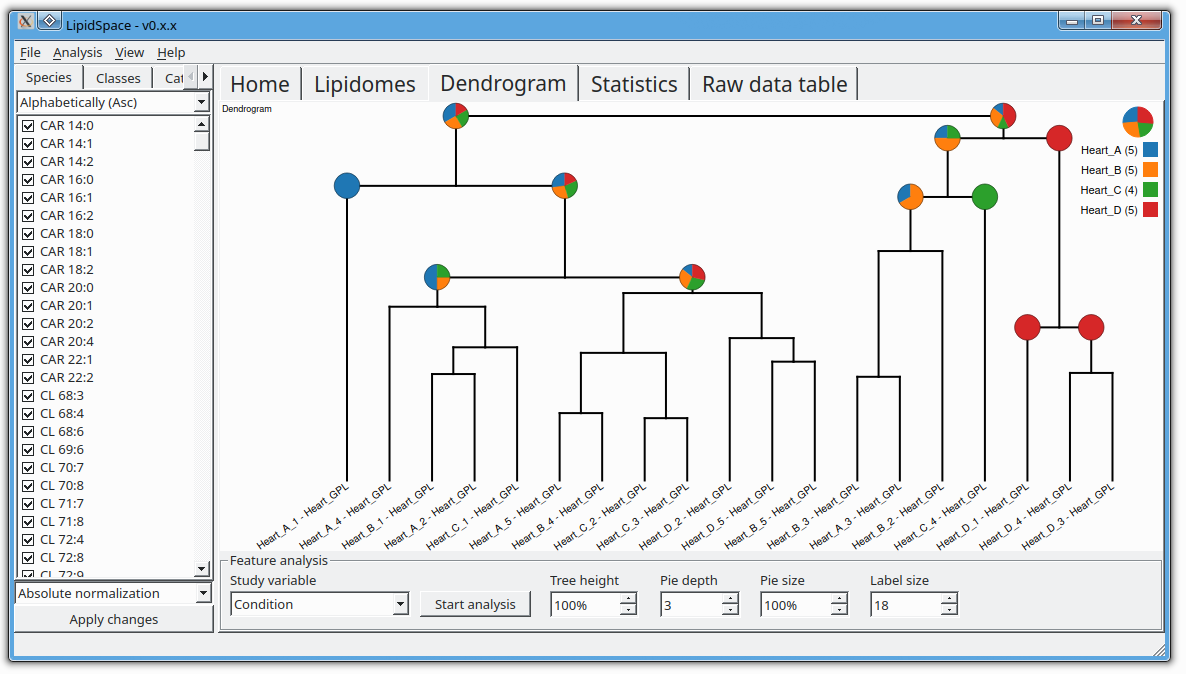


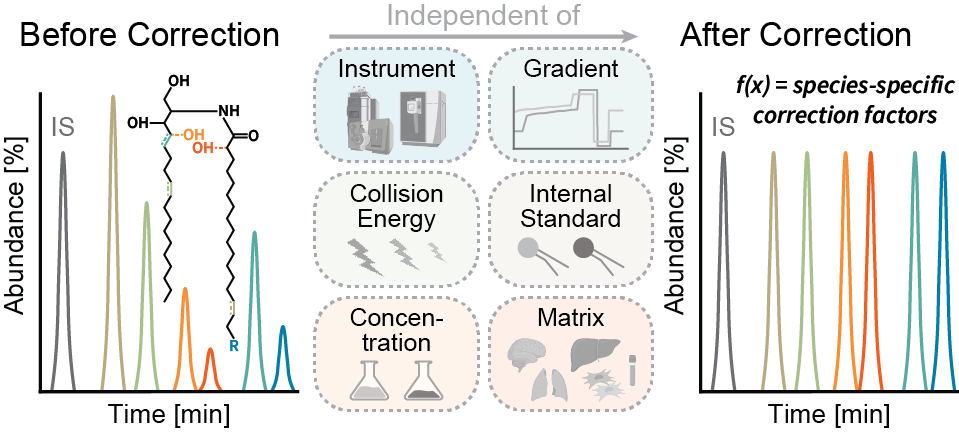




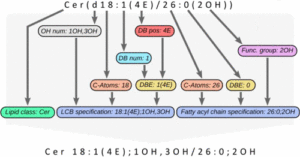 The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source.
The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source. 