
I was thrilled to have Gerhard Liebisch from Regensburg and Denise Wolrab with us on sample extraction. Both exciting talks gave our students great detail about what can go wrong in sample extraction during lipidomics experiments.
Gerhard Liebisch, Institute of Clinical Chemistry, Regensburg, Germany
PD Dr. Gerhard Liebisch obtained his Ph.D. at the University of Regensburg. His research interests focus on the development of MS methods for the quantification of lipid species. For more than 20 years, these methods have been applied in large-scale clinical studies and basic research. https://lipidomics-regensburg.de/

Denise Wolrab, University of Pardubice, Czech Republic
Dr. Denise Wolrab is an analytical chemist focusing on separation science and the application for the analysis of biomolecules, mainly using supercritical fluid chromatography hyphenated to mass spectrometry. After her Ph.D. at the University of Vienna in 2017, she obtained a postdoc position at the University of Pardubice responsible for the method development and application of lipidomics for cancer screening, mainly using UHPSFC/MS. Since 2020, Denise Wolrab has been PI and Grant Project manager at the University of Pardubice, evaluating the applicability of UHPSFC/MS for the metabolomic analysis of clinical samples.


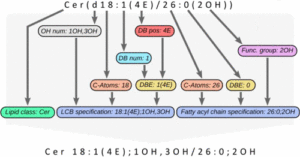 The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source.
The next version of GOSLIN is there! Goslin is the first grammar-based computational library for the recognition/parsing and normalization of lipid names following the hierarchical lipid shorthand nomenclature. The new version Goslin 2.0 implements the latest nomenclature and adds an additional grammar to recognize systematic IUPAC-IUB fatty acyl names as stored, e.g., in the LIPID MAPS database and is perfectly suited to update lipid names in LIPID MAPS or HMDB databases to the latest nomenclature. Goslin 2.0 is available as a standalone web application with a REST API as well as C++, C#, Java, Python 3, and R libraries. Importantly, it can be easily included in lipidomics tools and scripts providing direct access to translation functions. All implementations are open source. 




